Introducing pileup.js, a Browser-based Genome Viewer
19 Jun 2015Over the past few months, we’ve developed pileup.js, a new genome viewer designed for embedding within web applications. Try the demo!
While it’s still in the early stages of development, it can already load and visualize data from a variety of sources:
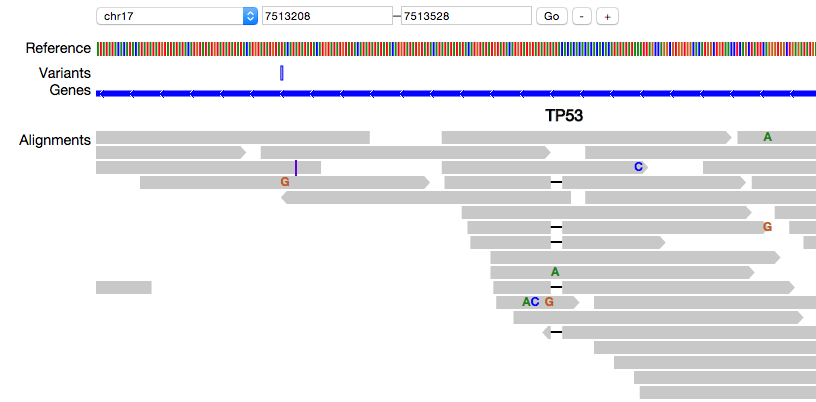
(pileup.js showing the reference genome, variants, genes and a pileup view of alignments)
pileup.js was inspired by two existing genome browsers: Biodalliance and IGV. Biodalliance is also designed to run in the browser, while IGV is a standalone Java desktop application (though the IGV team has recently begun work on a browser-based version, IGV.js).
pileup.js distinguishes itself from existing tools through the technical decisions that went into its design. The JavaScript ecosystem has grown tremendously over the past five years and creating a new genome browser from scratch has allowed us to take advantage of modern tooling:
- We use node-style modules via
requireand browserify. We pull in third-party dependencies via npm. - We use Mocha for testing and coveralls.io for tracking code coverage. We strive for very high test coverage: pileup.js is currently at 94%.
- We use Facebook’s FlowType type annotation system to do static analysis of our JavaScript. The public APIs are all well-annotated. Flow has helped us find bugs quickly and improve the readability of our code.
- We use a variety of ES6 features to make our code more concise, including arrow functions, template strings, classes and enhanced object literals. We transpile to browser-friendly JavaScript using jstransform.
In addition to modern tools, we’ve adopted several modern libraries:
- We make extensive use of Promises (via Q). Promises are a major improvement over the older callback style of asynchronous coding. They result in clearer control flow and greater composability.
- We use ReactJS to keep the various components of the pileup viewer in sync.
- We use D3 to render the visualizations in SVG layers.
By using SVG, we allow the tracks to be styled and positioned using CSS. This facilitates complex layouts and is more approachable for web developers than custom styling systems.
While pileup.js is being developed primarily for use within CycleDash, we expect that it will find uses in other web applications as well. The data fetching/parsing logic and the display are fully separated, so a web app could use pileup.js code to fetch BAM reads, for example, without rendering a visualization.
This is part of a broader goal of moving away from “bespoke” genomic file formats like BAM and VCF and towards entities which can be stored and served by non-filesystems, e.g. Impala or GA4GH.
You can learn more about pileup.js on GitHub or NPM.
Future areas for work include:
- Improving performance. While load times and frame rates while panning are acceptable today, we’d like to improve both in the future. This may require switching the rendering layer from SVG to canvas or WebGL.
- Adding more visualizations, for example read depth for alignment tracks or gene expression data.
- Factoring out the data-fetching code into a separate NPM package for easier reuse.
Please give pileup.js a try and let us know what you think. The best way to give feedback is through our issue tracker.