Introducing spark-bam
11 Dec 2017 | by Ryan WilliamsThe BAM file format is a common way of storing genomic reads that have been aligned to a reference genome. BAMs in the wild are frequently 10s to 100s of GBs of compressed data, and distributed/parallel processing is increasingly desirable.
Unfortunately, BAM is an “unsplittable” binary format; there’s no way of identifying record- or field-boundaries near arbitrary offsets within a BAM file without traversing all bytes from the start of the file.
Existing tools that process BAM files in distributed settings use hadoop-bam, an open-source library that implements heuristic-based “splitting” of BAM files. However, two problems have arisen as such tools have been more widely developed and deployed in disparate private- and public-cloud environments:
- correctness: record-boundary-identification heuristics emit “false positive” positions, resulting in attempts to read records from invalid data in the middle of records, crashing applications (or potentially silently corrupting analyses)
- performance: hadoop-bam computes record-boundary positions at fixed offsets (typically 64MB) on a single node, then uses all available nodes to process partitions delimited by those positions. On increasingly large BAMs – and data in public-cloud block-stores with long round-trip seeks – this split computation can take many minutes – costly wasted time while a whole cluster is potentially idling.
spark-bam addresses both of these issues, allowing fast, robust processing of BAM files with Apache Spark as well as tools for benchmarking its performance and correctness against hadoop-bam’s.
Correctness
spark-bam improves on hadoop-bam’s record-boundary-search heuristics in part by adding more checks:
| Validation check | spark-bam | hadoop-bam |
|---|---|---|
| Negative reference-contig idx | ✅ | ✅ |
| Reference-contig idx too large | ✅ | ✅ |
| Negative locus | ✅ | ✅ |
| Locus too large | ✅ | 🚫 |
Read-name ends with \0 |
✅ | ✅ |
| Read-name non-empty | ✅ | 🚫 |
| Invalid read-name chars | ✅ | 🚫 |
| Record length consistent w/ #{bases, cigar ops} | ✅ | ✅ |
| Cigar ops valid | ✅ | 🌓 |
| Subsequent reads valid | ✅ | ✅ |
| Non-empty cigar/seq in mapped reads | ✅ | 🚫 |
| Cigar consistent w/ seq len | 🚫 | 🚫 |
Correctness issues stemming from sensitivity to arbitrary internal buffer-sizes and JVM heap-size (!!) are also mitigated.
Benchmarking
spark-bam’s record-boundary-identification heuristics were tested against hadoop-bam’s on 1200+ BAMs from 4 publicly-available sequencing datasets totaling 20TB.
spark-bam was correct in all positions of all BAMs, while hadoop-bam’s accuracy varied widely across datasets:
- on short-read data with BAM records aligned to BGZF-block boundaries (e.g. selected BAMs from the 1000 Genomes Project), hadoop-bam is always correct
- on other short-read data (e.g. TCGA lung-cancer BAMs), hadoop-bam emits incorrect splits at rates between 1e-5 and 1e-6
- with 64MB splits, this corresponds to an expectation of one fault every 6-64TB of data processed
- in particular, the discovery of two TCGA BAMs where hadoop-bam crashes was the impetus for creating spark-bam
- on PacBio long read data (e.g. from the Genome in a Bottle project) the incorrect-split-rate is between 1e-4 and 1e-3
The spark-bam docs have more details about the experiments that were run and tools that run them.
Performance
hadoop-bam visits every partition boundary (typically every 64MB) in sequence before farming the adjusted partitions to cluster workers for parallel processing:
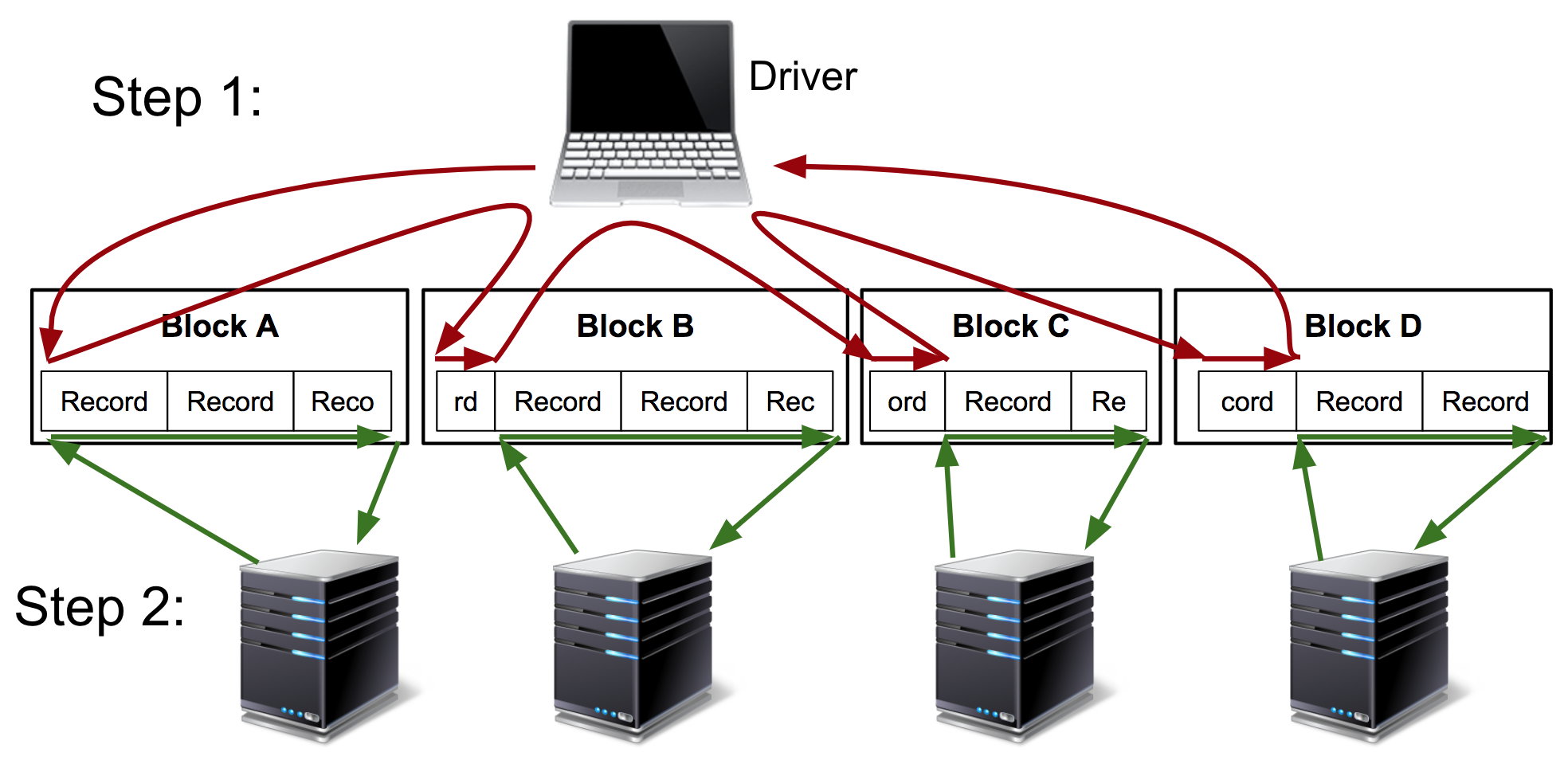
On the other hand, spark-bam pushes this partition-computation / “split-finding” to the workers: in a single Spark stage, each worker finds the first record-start position in its allotted range and processes records from that position to the end of its range:
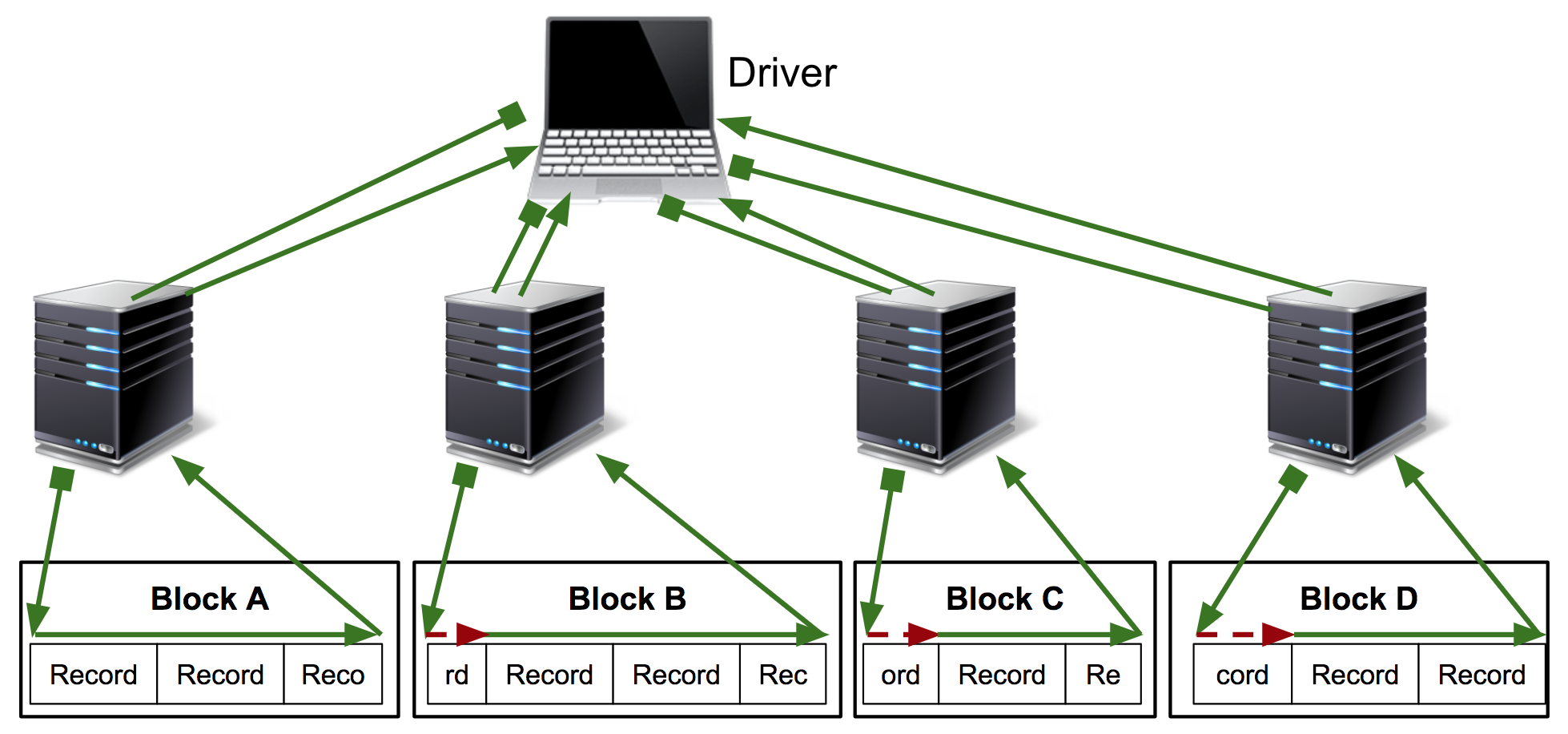
Depending on the number of partitions, number of workers, and network and disk-fetch times, this has been observed to result in speedups anywhere from “none” to several orders of magnitude.
CachingChannel
Another important optimization was a block-LRU-caching “channel” abstraction that mitigates pathological disk-access patterns while finding record boundaries.
To find a record boundary from an arbitrary position, the typical process is:
- attempt to read a record from current position
- ultimately determine that no valid record starts there, but having read several KB or MB ahead
- rewind to current position and advance by one byte
- try again from 1.
Short-read records are typically ≈300 bytes long, and underlying filesystem-IO libraries (like google-cloud-nio) might fetch/buffer 2MB at once, so on average finding one record boundary at the start of a 64MB partition might have involved fetching the same 2MB from disk a few hundred times in sequence.
Next steps
The spark-bam homepage has more information about using it, and the GitHub repo is open for issues and discussion.
Happy splitting!
